Nontuberculous Mycobacterial Pulmonary Disease
What are the criteria for diagnosing NTM pulmonary disease?
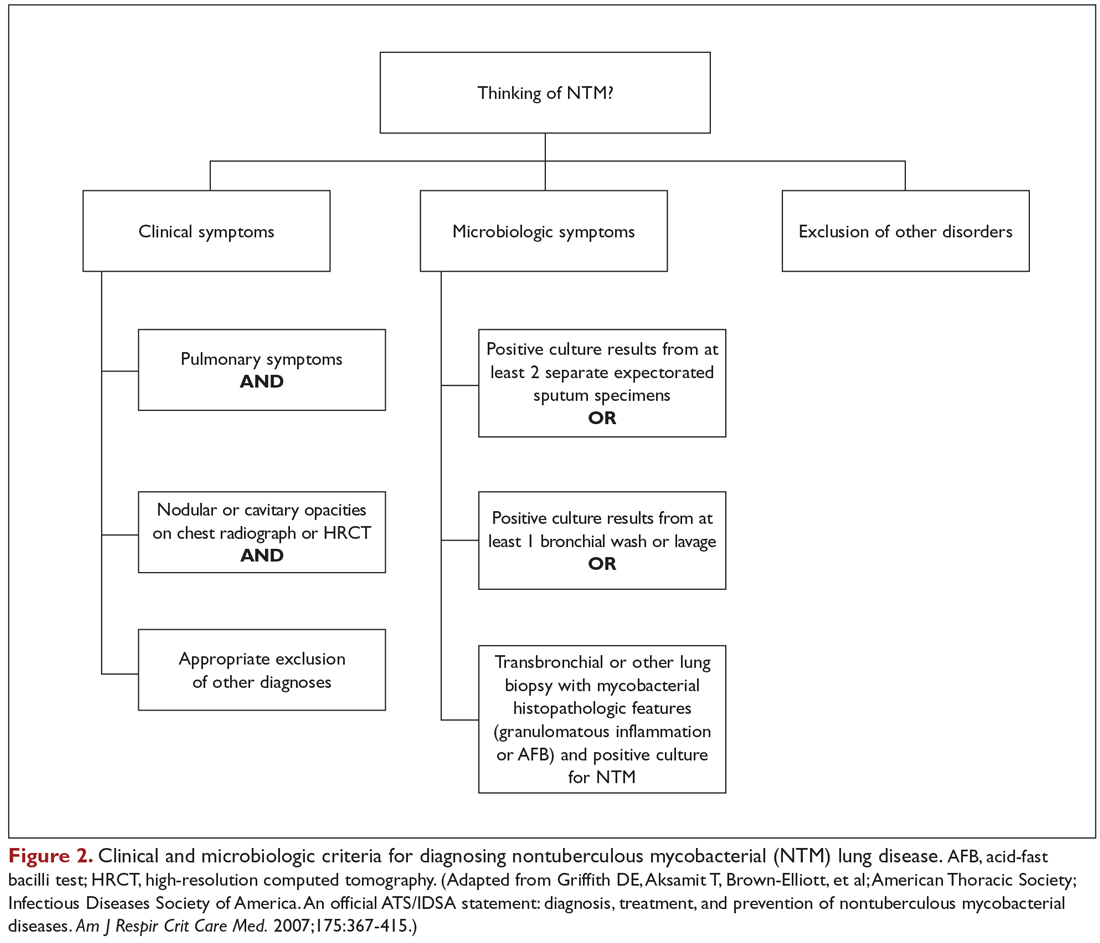
The diagnosis of NTM disease is based on clinical, radiologic, and mycobacterial correlation with good communication between the experts in this field. The ATS/IDSA criteria for diagnosing NTM lung disease are shown in Figure 2. These criteria best apply to MAC, M. kansasii, and M. abscessus, but are also clinically applied to other NTM respiratory pathogens. The diagnosis of MAC infection is most readily established by culture of blood, bone marrow, respiratory secretions/fluid, or tissue specimens from suspected sites of involvement. Due to erratic shedding of MAC into the respiratory secretions in patients with nodular bronchiectasis, as compared to those with the cavitary form of the disease, sputum may be intermittently positive, with variable colony counts and polyclonal infections.12 Prior to the advent of high-resolution CT, isolation of MAC organisms from the sputum of such patients was frequently dismissed as colonization.
Mycobacterial Testing
Because of the nonspecific symptoms and lack of diagnostic specificity of chest imaging, the diagnosis of NTM lung disease requires microbiologic confirmation. Specimens sent to the laboratory for identification of NTM must be handled with care to prevent contamination and false-positive results. Transport media and preservatives should be avoided, and transportation of the specimens should be prompt. These measures will prevent bacterial overgrowth. Furthermore, the yield of NTM may be affected if the patient has used antibiotics, such as macrolides and fluoroquinolones, prior to obtaining the specimen.
NTM should be identified at the species and subspecies level, although this is not practical in community practice settings. The preferred staining procedure in the laboratory is the fluorochrome method. Some species require special growth conditions and/or lower incubation temperatures, and other identification methods may have to be employed, such as DNA probes, polymerase chain reaction genotyping, nucleic acid sequence determination, and high-performance liquid chromatography. As a gold standard, clinical specimens for mycobacterial cultures should be inoculated onto 1 or more solid media (eg, Middlebrook 7H11 media and/or Lowenstein-Jensen media, the former of which is the preferred medium for NTM) and into a liquid medium (eg, BACTEC 12B broth or Mycobacteria growth indicator tube broth). Growth of visible colonies on solid media typically requires 2 to 4 weeks, but liquid media (eg, the radiometric BACTEC system), used as a supplementary and not as an exclusive test, usually produce results within 10 to 14 days. Furthermore, even after initial growth, identification of specific isolates based on the growth characteristics on solid media requires additional time. Use of specific nucleic acid probes for MAC and M. kansasii and HPLC testing of mycolic acid patterns in acid-fast bacilli smear–positive specimens can reduce the turnaround time of specific identification of a primary culture–positive sample. However, HPLC is not sufficient for definitive identification of many NTM species, including the RGM. Other newer techniques, including 16S ribosomal DNA sequencing and polymerase chain reaction-restriction fragment length polymorphism analysis, also allow NTM to be identified and speciated more reliably and rapidly from clinical specimens.
Cost and other practical considerations limit widespread adoption of these techniques. However, the recognition that M. abscessus can be separated into more than 1 subspecies, and that there are important prognostic implications of that separation, lends urgency to the broader adoption of newer molecular techniques in the mycobacteriology laboratory. Susceptibility testing is based on the broth microdilution method; RGM usually grow within 7 days of subculture, and the laboratory time to culture is a helpful hint, although not necessarily specific. Recognizing the morphology of mycobacterial colony growth may also be helpful in identification.
Are skin tests helpful in diagnosing NTM infection?
Tuberculin skin testing remains a nonspecific marker of mycobacterial infection and does not help in further elucidating NTM infection. However, epidemiologic and laboratory studies with well-characterized antigens have shown that dual skin testing with tuberculosis- versus NTM-derived tuberculin can discriminate between prior NTM and prior tuberculosis disease. Species-specific skin test antigens are not commercially available and are not helpful in the diagnosis of NTM disease because of cross-reactivity of M. tuberculosis and some NTM. However, increased prevalence of NTM sensitization based on purified protein derivative testing has been noted in a recent survey, which is consistent with an observed increase in the rates of NTM infections, specifically MAC, in the United States.26,27
Interferon-gamma release assays (IGRAs) are now being used as an alternative to tuberculin skin testing to diagnose M. tuberculosis infection. Certain NTM species also contain gene sequences that encode for ESAT-6 or CFP-10 antigens used in the IGRAs, and hence, yield a positive IGRA test. These include M. marinum, M. szulgai, and M. kansasii.28,29 However, MAC organisms do not produce positive results on assays that use these antigens.
Continue to: What is the approach to management of NTM pulmonary disease?






