The Diagnostic Yield of Noninvasive Microbiologic Sputum Sampling in a Cohort of Patients with Clinically Diagnosed Hospital-Acquired Pneumonia
The clinical predictors of positive sputum culture have not been previously reported in hospital-acquired pneumonia (HAP), and data on yield of sputum culture in this setting are scant. Current Infectious Disease Society of America guidelines for HAP recommend noninvasive sputum sampling, though the data for this practice are limited. We assessed the yield of sputum culture in HAP cases at an academic medical center from January 2007 to July 2013. HAP cases were identified by International Classification of Diseases, Ninth Revision-Clinical Modification codes for bacterial pneumonia and all cases were validated by chart review. Our cohort had 1172 hospitalizations with a HAP diagnosis. At least 1 sputum specimen was collected noninvasively and sent for bacterial culture after hospital day 2 and within 7 days of HAP diagnosis in 344 of these hospitalizations (29.4%), with a total of 478 sputum specimens, yielding 63 (13.2%) positive, 109 (22.8%) negative, and 306 (64.0%) contaminated cultures (>10 epithelial cells per high power field). Significant predictors of a positive sputum culture were chronic lung disease (relative risk [RR] = 2.0; 95% confidence interval [CI], 1.2-3.4) and steroid use (RR = 1.8; 95% CI, 1.1-3.2). The most commonly identified organisms were Gram-negative rods not further speciated (25.9%), Staphylococcus aureus (21.0%), and Pseudomonas aeruginosa (14.8%). Because of the ease of obtaining a sputum sample combined with the prevalence of commonly drug-resistant organisms, we suggest that sputum culture in HAP is a potentially useful noninvasive diagnostic technique.
© 2018 Society of Hospital Medicine
Microbiology
In our analysis, we used sputum samples obtained from expectorated or induced samples to evaluate the yield of noninvasive sputum sampling, as recommended by the IDSA. We included sputum samples collected ≥48 hours after admission and within 7 days of the clinical diagnosis of HAP. Sputum samples with >10 epithelial cells per high-power field (hpf) were considered to be contaminated. Among noncontaminated samples, positive sputum cultures were defined as those with a microbiologic diagnosis other than “oral flora,” while those with no growth or growth of oral flora or only yeast were considered to be negative. The hospital’s microbiology laboratory does not routinely provide species identification for Gram-negative rods (GNRs) growing on culture in the presence of growth of ≥3 other colony types. We considered such GNRs (not further speciated) to represent a positive culture result in our analysis given that colonization versus pathogenicity is a clinical distinction and, as such, these results may impact antibiotic choice.
Statistical Analysis
Data were analyzed by using SAS software, version 9.3. We used a 2-sided P value of <0.05 to indicate statistical significance for all comparisons. We used the χ2 test and the nonparametric median test for unadjusted comparisons.
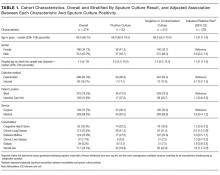
To identify predictors of a positive (versus negative or contaminated) sputum culture among patients with HAP, we used a generalized estimating equation model with a Poisson distribution error term, log link, and first-order autoregressive correlation structure to account for multiple sputum specimens per patient. We combined culture negative and contaminated samples to highlight the clinical utility of sputum culture in a real-world setting. Potential predictors chosen based on clinical grounds included all variables listed in Table 1. We defined comorbidities specified in Table 1 via ICD-9-CM secondary diagnosis codes and diagnosis related groups (DRGs) using Healthcare Cost and Utilization Project Comorbidity Software, version 3.7, based on the work of Elixhauser et al.9,10; dialysis use was defined by an ICD-9-CM procedure code of 39.95; inpatient steroid use was defined by a hospital pharmacy charge for a systemic steroid in the 7 days preceding the sputum sample.
RESULTS
There were 230,635 hospitalizations of patients ≥18 years of age from January 2007 to July 2013. After excluding outside hospital transfers (n = 14,422), hospitalizations <48 hours in duration (n = 59,774), and psychiatric hospitalizations (n = 9887), there were 146,552 hospitalizations in the cohort.
The top 3 bacterial organisms cultured from sputum samples were GNRs not further speciated (25.9%), Staphylococcus aureus (21.0%), and Pseudomonas aeruginosa (14.8%). The frequencies of isolated microorganisms are presented in Table 2.
In an adjusted analysis (Table 1), the significant predictors of a positive sputum culture were chronic lung disease (relative risk [RR] = 2.0; 95% confidence interval [CI], 1.2-3.4) and steroid use (RR = 1.8; 95% CI, 1.1-3.2).
DISCUSSION
To our knowledge, our study is the first to assess the predictors of positive sputum culture among patients with HAP (non-VAP) who had sputum samples obtained noninvasively, and this study is larger than prior studies in which researchers reported on sputum culture yield in HAP. Sputum samples were obtained in 29.4% cases of clinically diagnosed HAP. Although 87% of specimens obtained were culture-negative or contaminated, 13% yielded a bacterial organism. Although we do not report the antibiotic sensitivity patterns of the isolated organisms, the organisms identified frequently demonstrate antibiotic resistance, highlighting the potential for both antibiotic escalation and de-escalation based on sputum culture. In a multivariable model, presence of chronic lung disease and steroid use in the preceding week were both significantly associated with culture positivity.