Ordering and Interpreting Precision Oncology Studies for Adults With Advanced Solid Tumors: A Primer
Background: The promise of precision oncology can only be realized when genetic alterations are identified that can be leveraged to improve response and minimize toxicity. Identifying those alterations requires the knowledge to order the right test and to interpret the results correctly. This primer is designed to help clinicians order the appropriate testing for patients with specific malignancies and to give them an informed approach to interpretation.
Observations: Germline DNA is usually acquired from peripheral blood, buccal swab, or saliva collection in patients with a metastatic malignancy and can provide treatment options otherwise not available. However, germline testing does not indicate alterations that arise solely in tumor tissue. Somatic testing may be performed on primary tumor, metastatic biopsy, or circulating tumor DNA when the alteration is present at the time that the tumor developed and expected to be carried through the evolution of the tumor.
Conclusions: The rapid growth in technology and ability to enhance understanding of relevant tumor biology continues to improve the therapeutic landscape for individuals dealing with malignancy as does our ability to find targetable genetic alterations with the potential for meaningful clinical benefit.
Variant Interpretation of Germline Testing
A general understanding of the terminology used for germline variant interpretation allows for the ordering health care practitioner (HCP) to provide the best quality care and an appreciation for the limitations of current molecular testing. Not all variants are associated with disease; the clinical significance of a genetic variant falls on a spectrum. The criteria for determining pathogenicity differ between molecular laboratories, but most are influenced by the standards and guidelines set forth by the ACMG.14 The clinical molecular laboratory determines variant classification, and a detailed discussion is beyond the scope of this primer. In brief, variant classification is based on evidence of varying strength in different categories including population data, computational and predictive data, functional data, segregation data, de novo data, allelic data, and information from various databases. The ACMG has proposed a 5-tiered classification system, by which most molecular laboratories adhere to in their genetic test reports (Table 1).14
Pathogenic and likely pathogenic variants are clinically actionable, whereas variants of uncertain significance (VUS) require additional data and/or functional studies before making clinical decisions. Depending on the clinical context and existing supporting evidence, it may be prudent to continue monitoring for worsening or new signs of disease in patients with one or more VUS while additional efforts are underway to understand the variant’s significance.
In some cases, variants are reclassified, which may alter the management and treatment of patients. Reclassification can occur with VUS, and in rare instances, can also occur with variants previously classified as pathogenic/likely pathogenic or benign/likely benign. In such a case, the reporting laboratory will typically make concerted efforts to alert the ordering HCP. However, variant reclassifications are not always communicated to the care team. Thus, it is important to periodically contact the molecular laboratory of interest to obtain updated test interpretations.
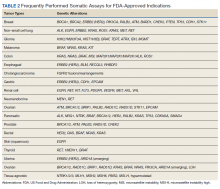
Somatic Testing
Testing of somatic (tumor) tissue is critical and is the approach most commonly taken in medical oncology (Table 2).
The advantages of primary tumor are that it is usually in hand as a diagnostic biopsy, acquisition is standard of care, and several targetable alterations are truncal, defined as driver mutations present at the time of tumor development. Also, the potential that the tumor arose in the background of a predisposing germline alteration can be suggested by sequencing primary tumor as discussed above. Moreover, sequencing the primary tumor can be done at any time unless the biopsy sample is considered too old or degraded (per specific platform requirements). The information gained can be used to anticipate additional treatment options that are relevant when patients experience disease progression. Disadvantages include the problem that primary specimens may be old or have limited tumor content, both of which increase the likelihood that sequencing will not be technically successful.
Alterations that are targetable and arise as a result of either treatment pressure or clonal evolution are considered evolutionary. If evolutionary alterations are the main focus for sequencing, then metastasis biopsy or ctDNA are better choices. The advantages of a metastasis biopsy are that tissue is contemporary, tumor content may be higher than in primary tumor, and both truncal and evolutionary alterations can be detected.
For specific tumors, continued analysis of evolving genomic alterations can play a critical role in management. In non–small cell lung cancer (NSCLC), somatic testing is conducted again at progression on repeat biopsies to evaluate for emerging resistance mutations. In epidermal growth factor receptor (EGFR)–mutated lung cancer, the resistance mutation, exon 20 p.T790M (point mutation), can present in patients after treatment with first- or second-generation EGFR tyrosine kinase inhibitors (TKI). Even in patients who are treated with the third-generation EGFR TKI osimertinib that can treat T790M-mutated lung cancer, multiple possible evolutionary mutations can occur at progression, including other EGFR mutations, MET/HER2 amplification, and BRAF V600E, to name a few.20 Resistance mechanisms develop due to treatment selection pressure and the molecular heterogeneity seen in lung cancer.
Disadvantages for metastatic biopsy include the inability to safely access a metastatic site, the time considerations for preauthorization and arrangement of biopsy, and a lower-than-average likelihood of successful sequencing from sites such as bone.21,22 In addition, there is some concern that a single metastatic site may not capture all relevant alterations for multiple reasons, including tumor heterogeneity.