Pharmacogenomic testing: Relevance in medical practice
ABSTRACTGenetics may account for much of the variability in our patients’ responses to drug therapies. This article offers the clinician an up-to-date overview of pharmacogenomic testing, discussing implications and limitations of emerging validated tests relevant to the use of warfarin (Coumadin), clopidogrel (Plavix), statins, tamoxifen (Nolvadex), codeine, and psychotropic drugs. It also discusses the future role of pharmacogenomic testing in medicine.
KEY POINTS
- Polymorphisms that affect the pharmacokinetics and pharmacodynamics of specific drugs are common.
- Testing for certain polymorphisms before prescribing certain drugs could help avoid adverse drug effects and improve efficacy.
- Pharmacogenomic testing has only recently begun to enter clinical practice, and routine testing is currently limited to a few clinical scenarios. However, additional applications may be just around the corner.
- Many pharmacogenomic tests are available, but testing has not yet been recommended for most drugs. Needed are large-scale trials to show that routine testing improves patient outcomes.
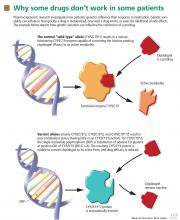
CLOPIDOGREL IS ACTIVATED BY CYP2C19
Clopidogrel, taken by about 40 million patients worldwide, is used to prevent atherothrombotic events and cardiac stent thrombosis when given along with aspirin.
Studies of clopidogrel pharmacogenomics
A recent genome-wide association study conducted in a cohort of 429 healthy Amish persons revealed a SNP in CYP2C19 to be associated with a diminished response to clopidogrel and to account for 12% of the variation in drug response.33 Traditional factors (the patient’s age, body-mass index, and cholesterol level) combined accounted for less than 10% of the variation.
Findings were similar in a subsequent investigation in 227 cardiac patients receiving clopidogrel: 21% of those with the variant had a cardiovascular ischemic event or died during a 1-year follow-up period compared with 10% of those without the variant (hazard ratio 2.42, P = .02).33
A 12-year prospective study investigating clopidogrel efficacy in 300 cardiac patients under the age of 45 used cardiovascular death, nonfatal myocardial infarction, and urgent coronary revascularization as end points. It concluded that the only independent predictor of these events was the patient’s CYP2C19 status.34
A study in 2,200 patients with recent myocardial infarction examined whether any of the known allelic variations that modulate clopidogrel’s absorption (ABCB1), metabolic activation (CYP3A4/5 and CYP2C19), or biologic activity (P2RY12 and ITGB3) was associated with a higher rate of the combined end point of all-cause mortality, nonfatal myocardial infarction, or stroke. None of the SNPs in CYP3A4/5, P2RY12, or ITGB3 that were evaluated was associated with a higher risk at 1 year. However, the allelic variations modulating clopidogrel’s absorption (ABCB1) and metabolism (CYP2C19) were associated with higher event rates. Patients with two variant ABCB1 alleles had a higher adjusted hazard ratio (95% confidence interval [CI] 1.2–2.47) than those with the wild-type allele. Patients who had one or two CYP2C19 loss-of-function alleles had a higher event rate than those with two wild-type alleles (95% CI 1.10–3.58 and 1.71–7.51, respectively).35
Conversely, researchers from the Population Health Research Institute found no association between poor-metabolizer status and treatment outcomes when CYP2C19 analysis was retrospectively added to the findings of two large clinical trials (combined N > 5,000). However, patients with acute coronary syndrome benefited more from clopidogrel treatment if they were ultra-rapid metabolizers (possessing the gain-of-function allele CYP2C19*17).36
Current status of clopidogrel testing: Uncertain
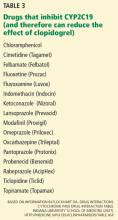
A current FDA boxed warning states that poor CYP2C19 metabolizers may not benefit from clopidogrel and recommends that prescribers consider alternative treatment for patients in this category.37 However, routine CYP2C19 testing is not recommended, and no firm recommendations have been established regarding dose adjustments for CYP2C19 status.
In 2010, the American College of Cardiology Foundation Task Force on Clinical Expert Consensus Documents and the American Heart Association collectively pronounced the current evidence base insufficient for recommending routine pharmacogenomic testing.39
Needed are large-scale studies examining the cost-effectiveness and clinical utility of genotype-guided clopidogrel therapy compared with other therapy options such as prasugrel (Effient), an analogue not metabolized by CYP2C19. One such study, sponsored by Medco Health Solutions, plans to enroll 14,600 cardiac patients and has an estimated completion date in June 2011.40 The expectation that clopidogrel will be available in generic form in 2012 adds to the uncertainty regarding the cost-effectiveness of CYP2C19 testing for clopidogrel therapy.
STATINS: SLC01B1*5 INCREASES MYOPATHY RISK
Statins lower the concentration of low-density lipoprotein cholesterol (LDL-C), resulting in a relative-risk reduction of about 20% for each 1 mmol/L (39 mg/dL) decrement in LDL-C.41 They are one of the most commonly prescribed classes of drugs, but their side effects can limit their appeal: statin-induced myopathy occurs in about 1:1,000 to 1:10,000 patients and is difficult to predict.
SLC01B1. The Study of the Effectiveness of Additional Reductions in Cholesterol and Homocysteine (SEARCH), a genome-wide association study, recently found a SNP (SLCO1B1* 5) in the SLC01B1 gene to be associated with a higher risk of statin-induced myopathy in cardiac patients receiving simvastatin (Zocor) 40 or 80 mg daily.42 The SLC01B1 gene, located on chromosome 12, influences the extent of the drug’s hepatic uptake and its serum concentration. Only the SLC01B1*5 SNP emerged as a predictor of statin-induced myopathy across the entire genome.42
The authors believe the findings are likely to apply to other statins. The mechanisms leading to statin-induced myopathy and the impact of statin pharmacogenomics are still unclear.43
CYP3A4. Other genetic variants may play a vital role in determining response to statin therapy. Carriers of a newly identified CYP3A4 polymorphism (intron 6 SNP, rs35599367, C>T) required significantly lower statin doses (0.2–0.6 times less) for optimal lipid control. The analyses included atorvastatin (Lipitor), simvastatin, and lovastatin (Mevacor), and the association was robust (P = .019).44
Statin pharmacogenomic testing is not routinely recommended
Routine pharmacogenomic testing for statin therapy is not recommended. Additional studies are needed to determine the clinical utility and cost-effectiveness of pharmacogenomic testing (involving a combination of several polymorphisms) in various patient populations delineated by type of statin, dose, and concomitant use of other drugs.